|
TinkerCell Core 1.0
TinkerCell's Core library providing all basic functionalities
|
|
TinkerCell Core 1.0
TinkerCell's Core library providing all basic functionalities
|
A set of node and connection families. All functions are static. More...
#include <Ontology.h>
Static Public Member Functions | |
| static NodeFamily * | nodeFamily (const QString &) |
| get a family pointer given its name (not case-sensitive) | |
| static ConnectionFamily * | connectionFamily (const QString &) |
| get a family pointer given its name (not case-sensitive) | |
| static bool | insertNodeFamily (const QString &, NodeFamily *) |
| add a new family to the ontology | |
| static bool | insertConnectionFamily (const QString &, ConnectionFamily *) |
| add a new family to the ontology | |
| static QList< NodeFamily * > | allNodeFamilies () |
| get list of all node families | |
| static QList< ConnectionFamily * > | allConnectionFamilies () |
| get list of all connection families | |
| static QStringList | allNodeFamilyNames () |
| get list of all node family names | |
| static QStringList | allConnectionFamilyNames () |
| get list of all connection family names | |
| static QStringList | readNodes (const QString &rdfFile, const QString &format=QString("rdfxml")) |
| read RDF file and insert node families | |
| static QStringList | readConnections (const QString &rdfFile, const QString &format=QString("rdfxml")) |
| read RDF file and insert connection families | |
| static void | cleanup () |
| delete family instances | |
A set of node and connection families. All functions are static.
Definition at line 40 of file Ontology.h.
| QList< ConnectionFamily * > Tinkercell::Ontology::allConnectionFamilies | ( | ) | [static] |
get list of all connection families
| QList<ConnectionFamily*> |
Definition at line 51 of file Ontology.cpp.
| QStringList Tinkercell::Ontology::allConnectionFamilyNames | ( | ) | [static] |
get list of all connection family names
| QStringList |
Definition at line 67 of file Ontology.cpp.
| QList< NodeFamily * > Tinkercell::Ontology::allNodeFamilies | ( | ) | [static] |
get list of all node families
| QList<NodeFamily*> |
Definition at line 43 of file Ontology.cpp.
| QStringList Tinkercell::Ontology::allNodeFamilyNames | ( | ) | [static] |
get list of all node family names
| QStringList |
Definition at line 59 of file Ontology.cpp.
| void Tinkercell::Ontology::cleanup | ( | ) | [static] |
delete family instances
Definition at line 413 of file Ontology.cpp.
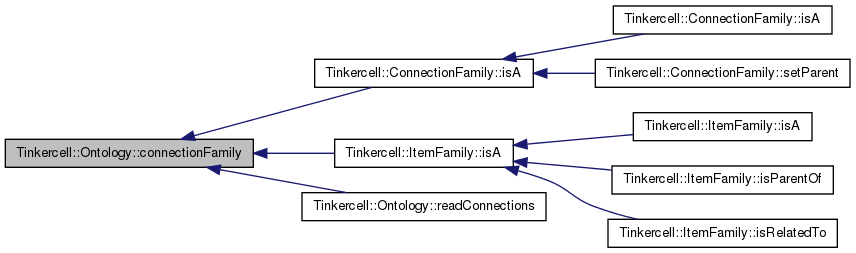
| ConnectionFamily * Tinkercell::Ontology::connectionFamily | ( | const QString & | s0 | ) | [static] |
get a family pointer given its name (not case-sensitive)
| QString& | name of family |
Definition at line 21 of file Ontology.cpp.
| bool Tinkercell::Ontology::insertConnectionFamily | ( | const QString & | s0, |
| ConnectionFamily * | ptr | ||
| ) | [static] |
add a new family to the ontology
| QString | family name |
| ConnectionFamily* | new family |
Definition at line 36 of file Ontology.cpp.
| bool Tinkercell::Ontology::insertNodeFamily | ( | const QString & | s0, |
| NodeFamily * | ptr | ||
| ) | [static] |
add a new family to the ontology
| QString | family name |
| NodeFamily* | new family |
Definition at line 29 of file Ontology.cpp.
| NodeFamily * Tinkercell::Ontology::nodeFamily | ( | const QString & | s0 | ) | [static] |
get a family pointer given its name (not case-sensitive)
| QString& | name of family |
Definition at line 13 of file Ontology.cpp.
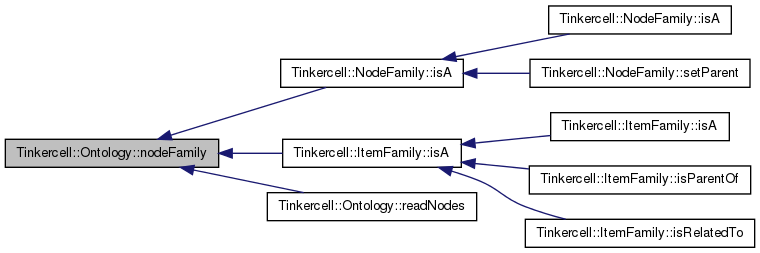
| QStringList Tinkercell::Ontology::readConnections | ( | const QString & | rdfFile, |
| const QString & | format = QString("rdfxml") |
||
| ) | [static] |
read RDF file and insert connection families
| QString | file name |
| QString | format, defaults to rdfxml |
Definition at line 359 of file Ontology.cpp.
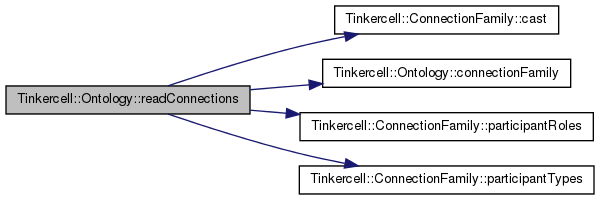
| QStringList Tinkercell::Ontology::readNodes | ( | const QString & | rdfFile, |
| const QString & | format = QString("rdfxml") |
||
| ) | [static] |
read RDF file and insert node families
| QString | file name |
| QString | format, defaults to rdfxml |
Definition at line 322 of file Ontology.cpp.
 1.7.4
1.7.4